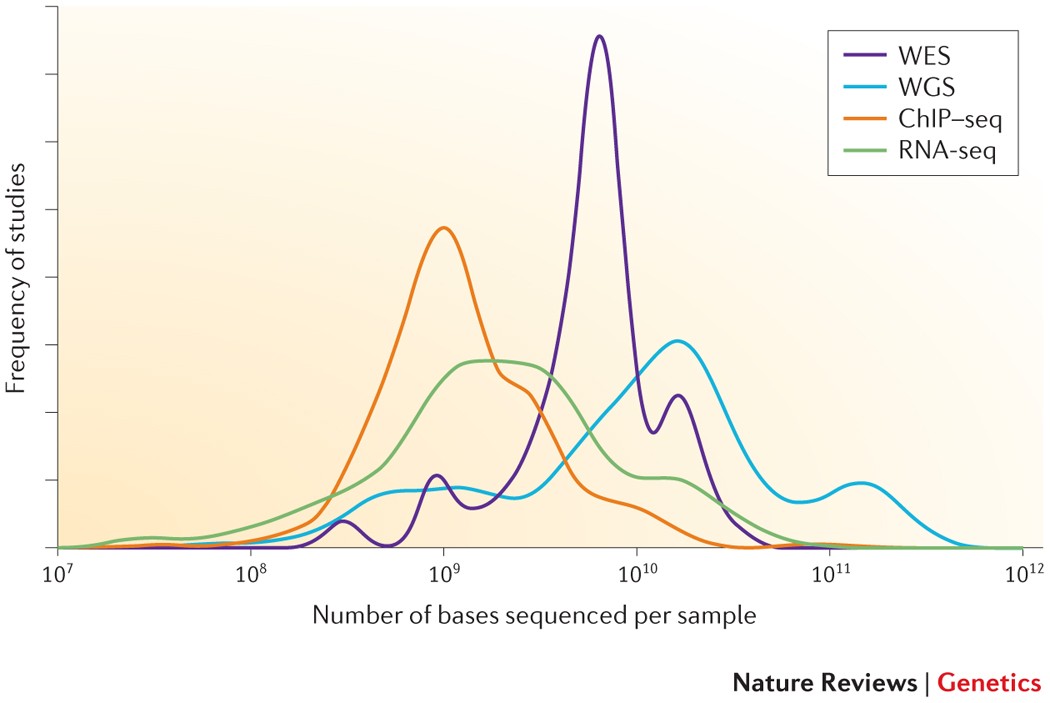
Detecting, Categorizing, and Correcting Coverage Anomalies of RNA-Seq Quantification - ScienceDirect
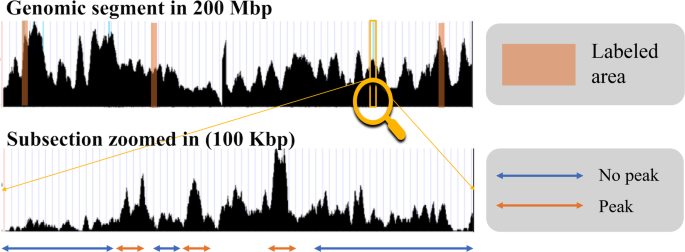
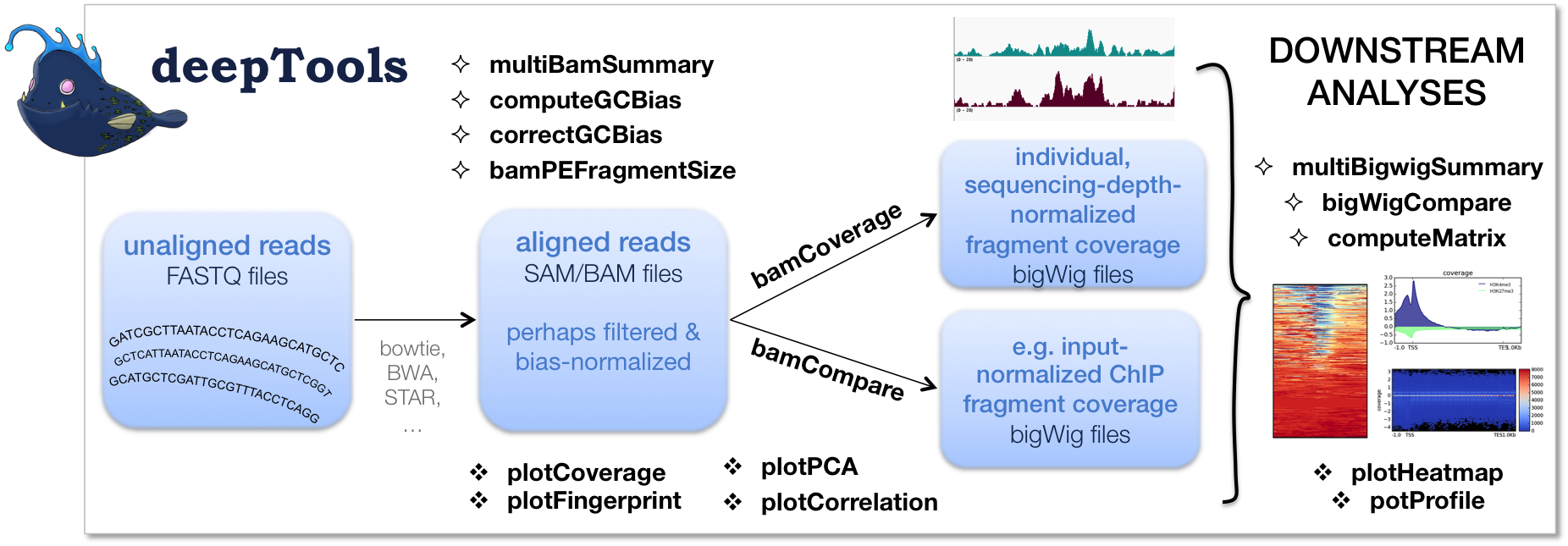
CNN-Peaks: ChIP-Seq peak detection pipeline using convolutional neural networks that imitate human visual inspection | Scientific Reports

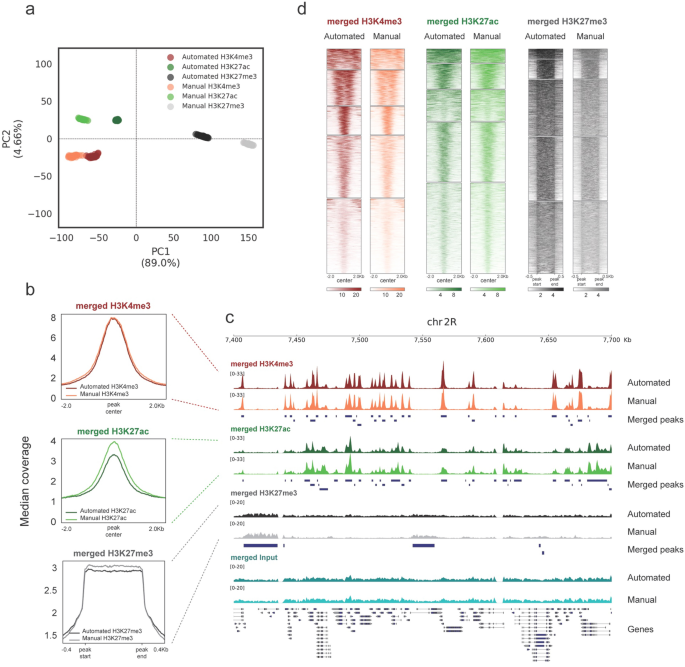
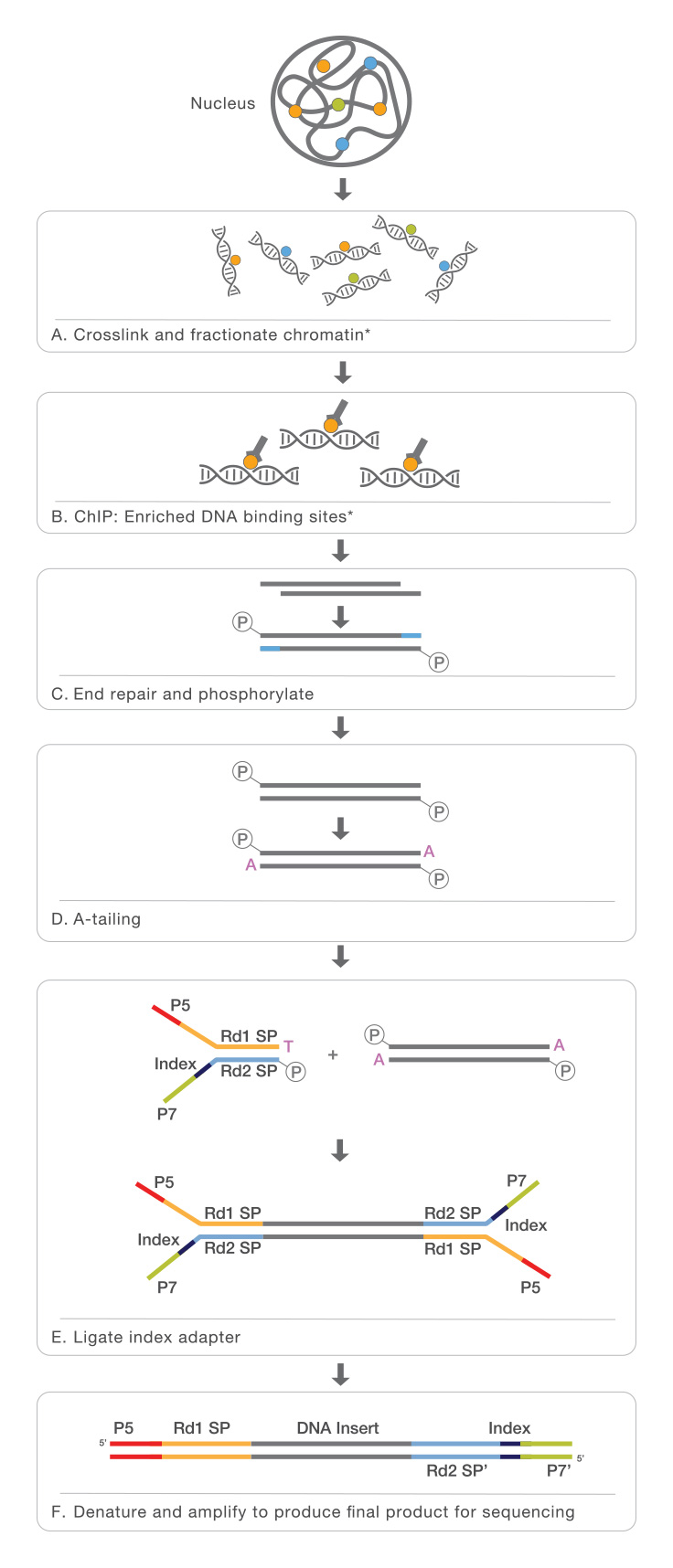
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance

ChIP-Seq deconvolution maps are highly reproducible. (A) Comparison of... | Download Scientific Diagram
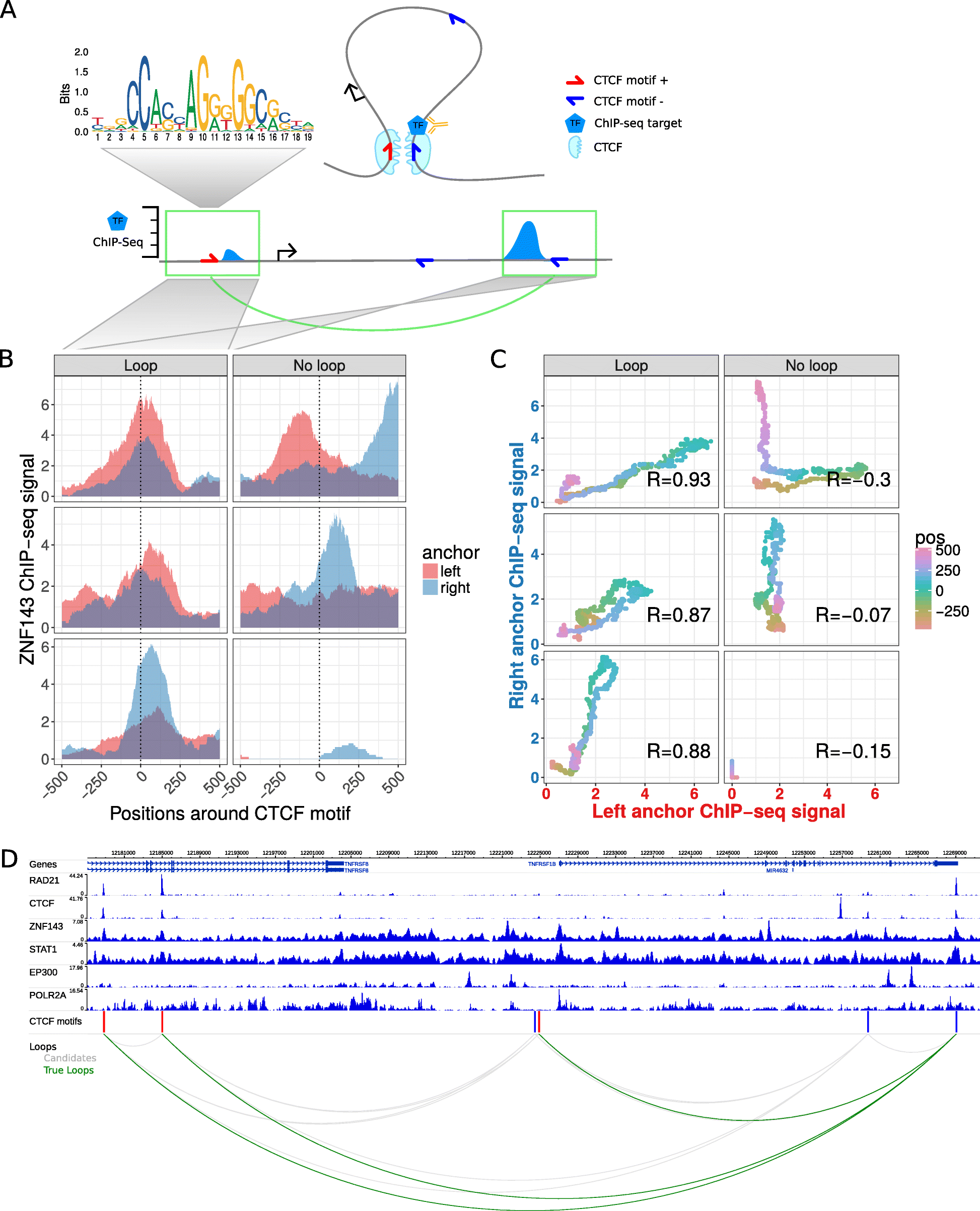
7C: Computational Chromosome Conformation Capture by Correlation of ChIP-seq at CTCF motifs | BMC Genomics | Full Text

Example genome browser screen-shot. ChIP-seq read coverage of H3K27me3... | Download Scientific Diagram

ChIP-seq peak types from various experiments.(a–c) Data shown are from... | Download Scientific Diagram

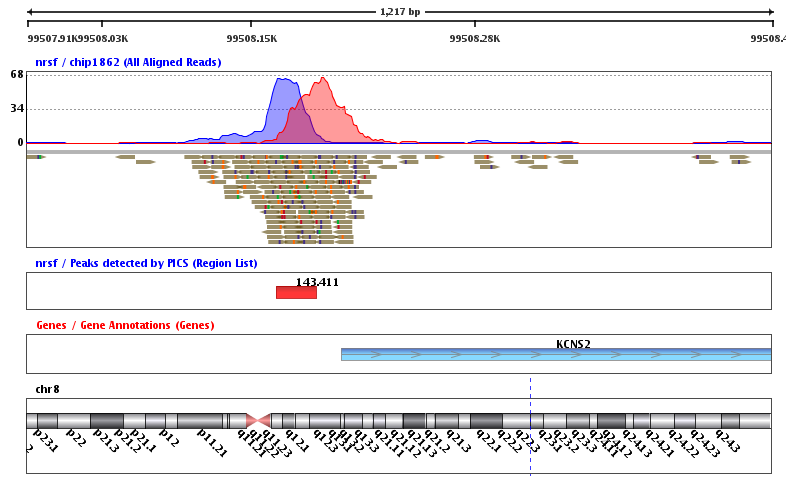
Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II | PNAS